Modules
GenomeRunner is installed as a grsnp module. There are four submodules in the package:
dbcreator_ucscanddbcreator_encodeDCC- modules for creating organism-specific genome annotation databasesoptimizer- a module to speed up GenomeRunner by pre-calculating background overlap statisticsserver- a module for the web interfacehypergeom4- a command-line module for the enrichment/epigenomic similarity analyses
The interplay among the modules is shown on the figure
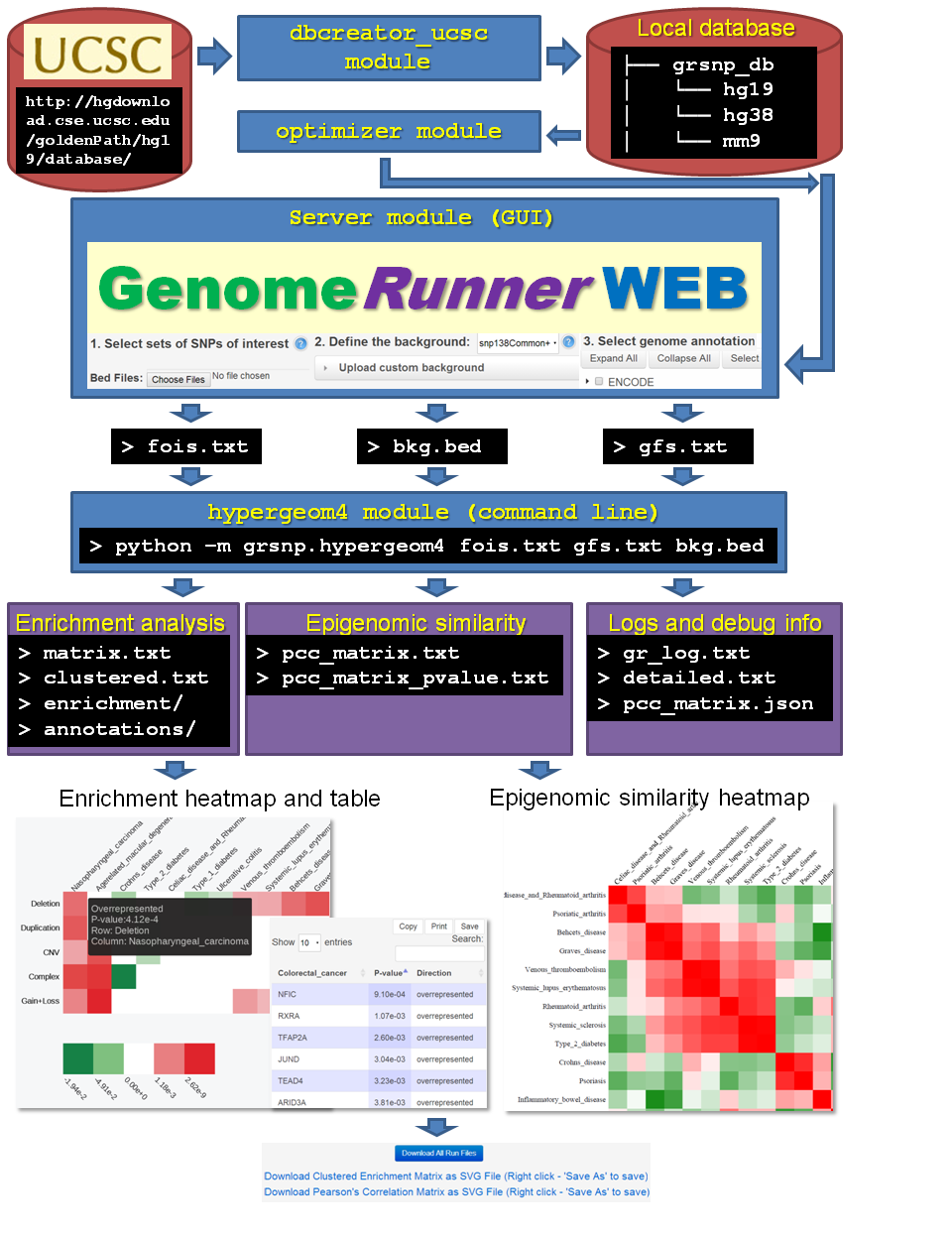
Before running GenomeRunner, the first step is to create a database of genomic annotations using the dbcreator module. Optionally, optimize the database using the optimizer module. Then, run GenomeRunner web server using the server module. To run GenomeRunner from command line, use the hypergeom4 module.